phaseportnormal2.mws
Plot phase portrait for moment closure equations for Verhulst logistic model, assuming normal distribution.
This Maple work sheet uses Maple8, and the new package LinearAlgebra. Date 030402.
| > | restart; with(DEtools,DEplot); with(plots,display); with(LinearAlgebra,Eigenvectors); with(VectorCalculus,Jacobian); XP:=Vector(2); |
![]()
![]()
![]()
![]()
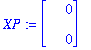
In the original Verhulst model, take alpha1=alpha2=1, R0=4, N=100. Then a1=3/5, f1=5/3, f2=3/5. Take r/K=1 (by rescaling time). Put kappa1=X[1], kappa2=X[2].
The equations are given by the procedure G:
| > | G:=proc(N,t,X,XP) XP[1]:=(60-X[1])*X[1]-X[2]; XP[2]:=(100-0.6*X[1])*X[1]+119.4*X[2]-4*X[1]*X[2]; end; |
![]()
![]()
![]()
Run G to determine XP:
| > | G(2,t,X,XP): |
We determine the isoclines where the derivatives are equal to zero:
| > | isocline1:=solve(XP[1],X[2]); isocline2:=solve(XP[2],X[2]); |
![]()
![isocline2 := -1.*X[1]*(-500.+3.*X[1])/(-597.+20.*X[1])](images/phaseportnormal210.gif)
Plot the isoclines:
| > | plot({isocline1,isocline2},X[1]=-10..100,y=-100..1000,discont=true); |
![[Maple Plot]](images/phaseportnormal211.gif)
| > | expand(subs(X[2]=isocline1,XP[2])); |
![]()
| > | plot(%,X[1]=-10..70); |
![[Maple Plot]](images/phaseportnormal213.gif)
There are three critical points. Solve for them:
| > | critp:=solve({XP[1],XP[2]},{X[1],X[2]}); |
![]()
![]()
I use P1, P2, P3 to denote the three critical points. They are numbered from the left.
CHECK the numbering here!
| > | P1prel:=critp[1]; P2prel:=critp[2]; P3prel:=critp[3]; |
![]()
![]()
![]()
CHECK the ordering here!
| > | P1:=[rhs(P1prel[1]),rhs(P1prel[2])]; P2:=[rhs(P2prel[1]),rhs(P2prel[2])]; P3:=[rhs(P3prel[1]),rhs(P3prel[2])]; |
![]()
![]()
![]()
Now determine the Jacobian of the right-hand sides of the given system of equations:
| > | J:=Jacobian(XP,[X[1],X[2]]); |
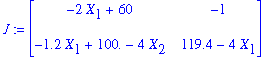
Determine the jacobian in each of the critical points:
| > | J1:=subs(P1prel,J); |
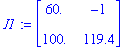
| > | J2:=subs(P2prel,J); |
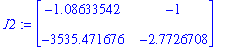
| > | J3:=subs(P3prel,J); |
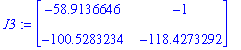
The eigenvalues and eigenvectors of these three matrices:
| > | eg1:=Eigenvectors(J1,output=list); |
| > | eg2:=Eigenvectors(J2,output=list); |
| > | eg3:=Eigenvectors(J3,output=list); |
![eg1 := [[61.7341279413639583+0.*I, 1, Vector(%id = 5754832)], [117.665872058636040+0.*I, 1, Vector(%id = 5754872)]]](images/phaseportnormal226.gif)
![eg1 := [[61.7341279413639583+0.*I, 1, Vector(%id = 5754832)], [117.665872058636040+0.*I, 1, Vector(%id = 5754872)]]](images/phaseportnormal227.gif)
![eg2 := [[57.5363073002976151+0.*I, 1, Vector(%id = 6017656)], [-61.3953135202976128+0.*I, 1, Vector(%id = 5996020)]]](images/phaseportnormal228.gif)
![eg2 := [[57.5363073002976151+0.*I, 1, Vector(%id = 6017656)], [-61.3953135202976128+0.*I, 1, Vector(%id = 5996020)]]](images/phaseportnormal229.gif)
![eg3 := [[-57.2699014927418233+0.*I, 1, Vector(%id = 5755272)], [-120.071092307258183+0.*I, 1, Vector(%id = 6114996)]]](images/phaseportnormal230.gif)
![eg3 := [[-57.2699014927418233+0.*I, 1, Vector(%id = 5755272)], [-120.071092307258183+0.*I, 1, Vector(%id = 6114996)]]](images/phaseportnormal231.gif)
Notice that P1 is an unstable node, P2 is a saddle point, P3 is a stable node. Furthermore,
CHECK the numbering here!
eg1[1] : slow manifold, eg1[2]: fast manifold of unstable node P0. Both manifolds have negative slopes.
eg2[1]: unstable manifold, eg2[2]: stable manifold of saddle point P1.
eg3[1]: slow manifold, eg3[2]: fast manifold of stable node P2.
A Maple procedure for determining initial values on each manifold near each critical point...
Think of it as transforming eg-to-init (eg2init):
| > | eg2init:=proc(P,eg,d) local u,lu,v,k; u:=eg[3]; # This is an eigenvector. lu:=sqrt(u[1]^2+u[2]^2); # This is the length of the eigenvector u. v:=d*u/lu; # This is a vector of length d in the direction of u. [seq([x(0)=P[1]+(-1)^k*v[1],y(0)=P[2]+(-1)^k*v[2]],k=1..2)]; #These are two initial values near P in the direction of the eigenvector. end proc: |
CHECK the numbering here!
| > | init2unstable:=eg2init(P2,eg2[1],10); init2stable:=eg2init(P2,eg2[2],10); init3fast:=eg2init(P3,eg3[2],10); |
![]()
![]()
![]()
![]()
![]()
![]()
Plot orbits on the unstable manifold at P2:
| > | plot2unstable:=DEplot(G,[x(t),y(t)],t=0..0.2,number=2,init2unstable,x=-10..100,y=-100..1000,arrows=none,stepsize=0.01,linecolor=red,thickness=4): plot2unstable; |
![[Maple Plot]](images/phaseportnormal238.gif)
Plot orbits on the stable manifold at P2:
| > | plot2stable:=DEplot(G,[x(t),y(t)],t=-0.2..0,number=2,init2stable,x=-10..100,y=-100..1000,arrows=none,stepsize=0.01,linecolor=red,thickness=4): plot2stable; |
![[Maple Plot]](images/phaseportnormal239.gif)
Plot orbits on the fast manifold at P3:
| > | plot3fast:=DEplot(G,[x(t),y(t)],t=-0.04..0,number=2,init3fast,x=-10..100,y=-100..1000,arrows=none,linecolor=red,stepsize=0.01,thickness=4): plot3fast; |
![[Maple Plot]](images/phaseportnormal240.gif)
Plot one orbit on the slow manifold at P3:
| > | plot3slow:=DEplot(G,[x(t),y(t)],t=0..0.1,number=2,{[x(0)=500,y(0)=20]},x=-10..100,y=-100..1000,stepsize=0.002,arrows=none,linecolor=red,thickness=4): plot3slow; |
![[Maple Plot]](images/phaseportnormal241.gif)
NOTE: The arrows on the final phase portrait become less pronounced if the commend "color=red" below is replaced by "color=yellow".
Plot a direction field:
| > | plotarrows:=DEplot(G,[x(t),y(t)],t=0..0.01,number=2,{[x(0)=60,y(0)=32]},x=-10..100,y=-100..1000,stepsize=0.01,color=red,linecolor=red): plotarrows; |
![[Maple Plot]](images/phaseportnormal242.gif)
Plot three additional orbits:
| > | init1:=[x(0)=10,y(0)=0]; plot1:=DEplot(G,[x(t),y(t)],t=0..0.1,number=2,[init1],x=-10..100,y=-100..1000,stepsize=0.01,arrows=none,linecolor=red,thickness=4): plot1; |
![]()
![[Maple Plot]](images/phaseportnormal244.gif)
| > | init2:=[x(0)=50,y(0)=1000]; plot2:=DEplot(G,[x(t),y(t)],t=0..0.1,number=2,[init2],x=-10..100,y=-100..1000,stepsize=0.01,arrows=none,linecolor=red,thickness=4): plot2; |
![]()
![[Maple Plot]](images/phaseportnormal246.gif)
| > | init3:=[x(0)=100,y(0)=1000]; plot3:=DEplot(G,[x(t),y(t)],t=0..0.1,number=2,[init3],x=-10..100,y=-100..1000,stepsize=0.002,arrows=none,linecolor=red,thickness=4): plot3; |
![]()
![[Maple Plot]](images/phaseportnormal248.gif)
Plot all these orbits in one figure:
| > | display({plot2stable,plot2unstable,plot3fast,plot3slow,plotarrows,plot1,plot2,plot3},labels=[Mean,Variance],title=Normal_Assumption); |
![[Maple Plot]](images/phaseportnormal249.gif)
| > |